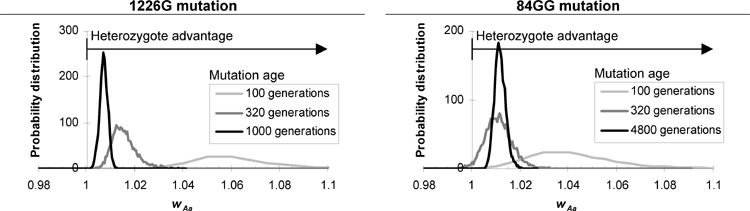
Based on linkage disequilibrium, we show that the 1226G Gaucher disease mutation probably originated less than 1000 generations ago. Its recent origin and high current allele frequency provide strong evidence for heterozygote selection.

Furthermore, we show that the linkage disequilibrium between a mutation and a nearby marker can serve as a molecular clock for the age of the mutation, but only after accounting for some of the clock's peculiar properties. For example, this "clock" can easily get stuck at zero in small populations or closely linked genes due to random loss of recombinant chromosomes. This paper's analysis of the variations in the clock speed allows us to better interpret linkage data.
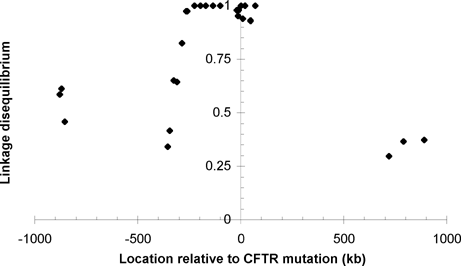
An interesting corollary of this analysis is that markers extremely close to a locus of interest may stay more tightly linked to it than the expected exponential decay of linkage disequilibrium with distance. This phenomenon has been observed with markers near the ΔF508 mutation of the cystic fibrosis transmembrane conductance regulator (CFTR) gene.
|
View the:
Paper - © 2000 by Academic Press. Publisher's note: "This material has been published in Blood Cells, Molecules, and Diseases, August 2000, 26(4): 348-59, the only definitive repository of the content that has been certified and accepted after peer review. Copyright and all rights therein are retained by Academic Press" |
Boas, F. Edward. (2000) "Linkage to Gaucher Mutations in the Ashkenazi Population: Effect of Drift on Decay of Linkage
Disequilibrium and Evidence for Heterozygote Selection." Blood Cells, Molecules, and Diseases. 26(4): 348-59.
Abstract |